DISTEVAL: Protein Distance Evaluation
DISTEVAL: A web-server for evaluating predicted protein distances
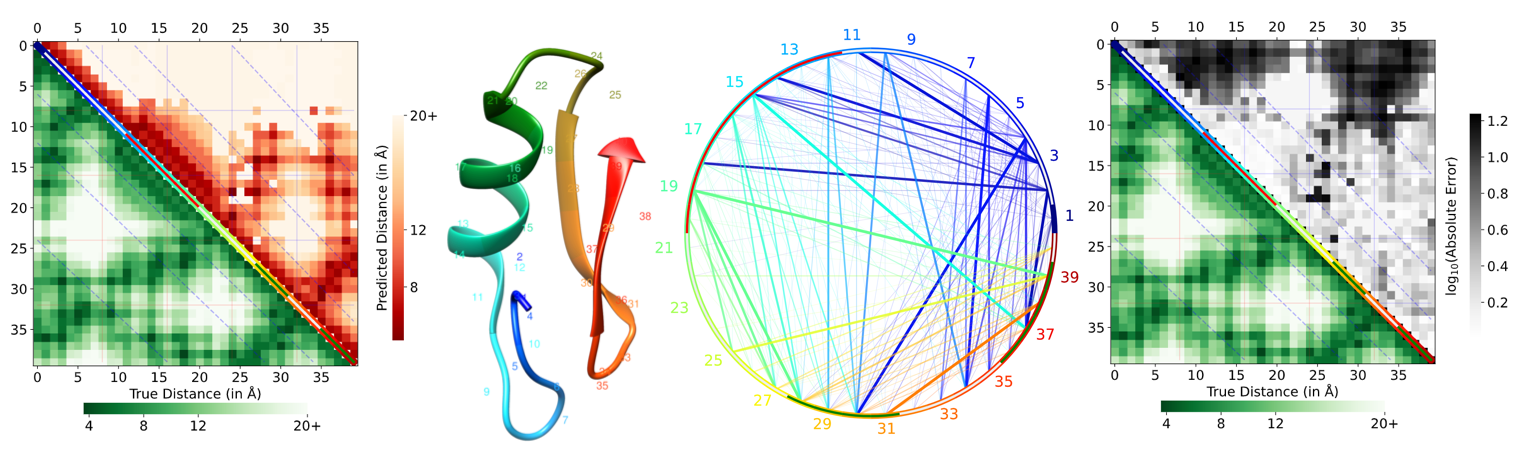
Predicted protein inter-residue contacts and distances are the key intermediate steps towards accurate protein structure prediction. Distance prediction or distance-range prediction is a more granular version of the contact prediction problem and it is now introduced as a new challenge in the CASP14 experiment. Despite the recent proliferation of methods for predicting distances, no methods currently exist for evaluating predicted distances. This work discusses a new web-server for evaluating predicted protein inter-residue distances. The server accepts predicted contacts or distances along with a true structure as input. It generates informative chord diagrams and heat maps to facilitate visual assessment and evaluates predictions using MAE and the standard ‘contact precision’ metric. Our tool, DISTEVAL, is available at http://deep.cs.umsl.edu/disteval/.